2. Shell scripts for independent contrast regression analysis
Source:vignettes/articles/phyreg_shell.Rmd
phyreg_shell.Rmd
The main goal of the InNOutBT package is to readily create input data and process output data for Meade & Pagel’s (2022) BayesTraits program.
Here in this article, we show how to use InNOutBT’s function phyreg.shell for producing shell scripts and associated run commands of BayesTraits phylogenetic independent contrast regression analyses. Basically, the function phyreg.shell uses the directory paths to the tree, mean and linked data files as created with phyreg.inputs (see article here), to automatically produce shell scripts in both Unix and Windows PowerShell formats. The resulting single shell script file can then be used for running one or all-in-once multiple BayesTraits phylogenetic regressions, without manually setting new analyses with new parameters. The shell script produced with phyreg.shell should have thus all parameters for all analyses with any defined model. In addition to save the shell script directly in BayesTraits directory, the function saves all the separate running files with BayesTraits specified parameters (bi, it, sa, and st) for any run, depending on the chosen model of tree transformation. See BayesTraits V4.0.0 Manual for further details on each of BayesTraits parameters available for phylogenetic regression so as to better understand how to set the arguments of phyreg.shell.
Setup
InNOutBT is not on CRAN yet but you can install the latest development version from GitHub:
#install.packages("devtools")
devtools::install_github("DBOSlab/InNOutBT")Creating Unix shell script
Set the argument syst = "unix" to create a shell script for running the phylogenetic regressions with macOS Monterey.
Note that you should give the name of the response variable of your analysis in the argument responvar. Also, you should use the argument treetransf to define any or all (listed as a vector) available tree transformation models that you want to run the phylogenetic regressions with your input data. As such, choose among the models Kappa, Delta, Lambda, OU (Ornstein–Uhlenbeck), UNI (if you have set a complete default BayesTraits regression analysis, i.e. assumed a uniform rates of evolution no other tree transformation models were estimated), VR (if you have used a variable rates) or Fabric (if you want to use the general statistical mode that accommodates uneven evolutionary landscape as described by Pagel et al. 2022).
library(InNOutBT)
phyreg.shell(meanfile_dir = "results_BayesTraits_phyreg_input/09Jun2022",
linkfile_dir = "results_BayesTraits_phyreg_input/09Jun2022",
treefile_dir = "Data",
BayesTraits_dir = "/Users/domingoscardoso/BayesTraitsV4",
responvar = "DBH",
treetransf = c("UNI", "Lambda", "VR", "Fabric"),
Kappa = NULL,
Delta = NULL,
Lambda = NULL,
OU = NULL,
VRLambda = FALSE,
bi = "30000000",
it = "130000000",
sa = "100000",
st = c("500", "50000"),
syst = "unix",
dir_create = "results_BayesTraits_phyreg_shell",
cc_DataTree = TRUE)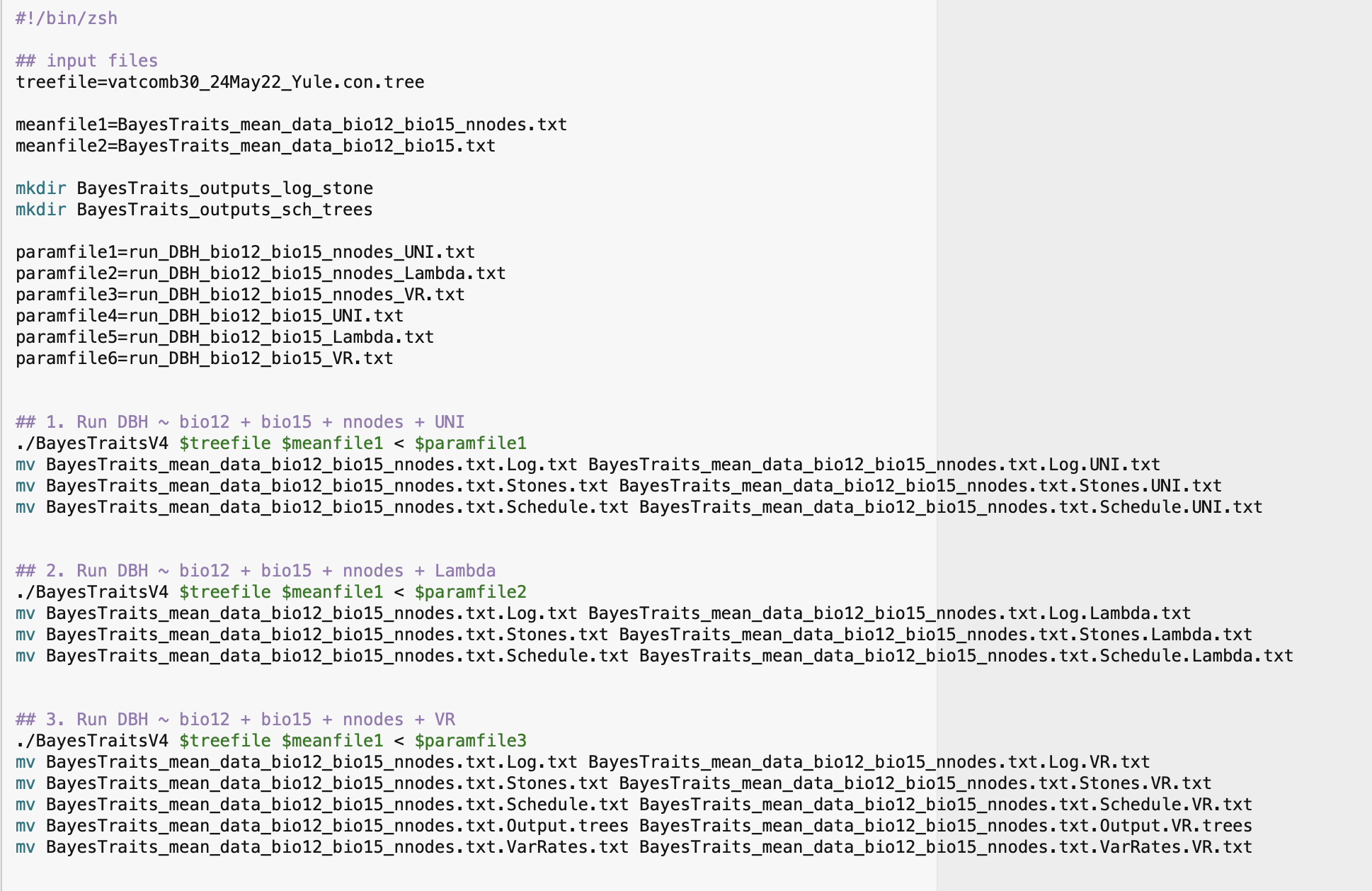
Creating Windows PowerShell script
Set the argument syst = "windows" to create a PowerShell script for running the phylogenetic regressions with Windows.
phyreg.shell(meanfile_dir = "results_BayesTraits_phyreg_input/09Jun2022",
linkfile_dir = "results_BayesTraits_phyreg_input/09Jun2022",
treefile_dir = "Data",
BayesTraits_dir = "/Users/domingoscardoso/BayesTraitsV4",
responvar = "DBH",
treetransf = c("UNI", "Lambda", "VR", "Fabric"),
Kappa = NULL,
Delta = NULL,
Lambda = NULL,
OU = NULL,
VRLambda = FALSE,
bi = "30000000",
it = "130000000",
sa = "100000",
st = c("500", "50000"),
syst = "windows",
dir_create = "results_BayesTraits_phyreg_shell",
cc_DataTree = TRUE)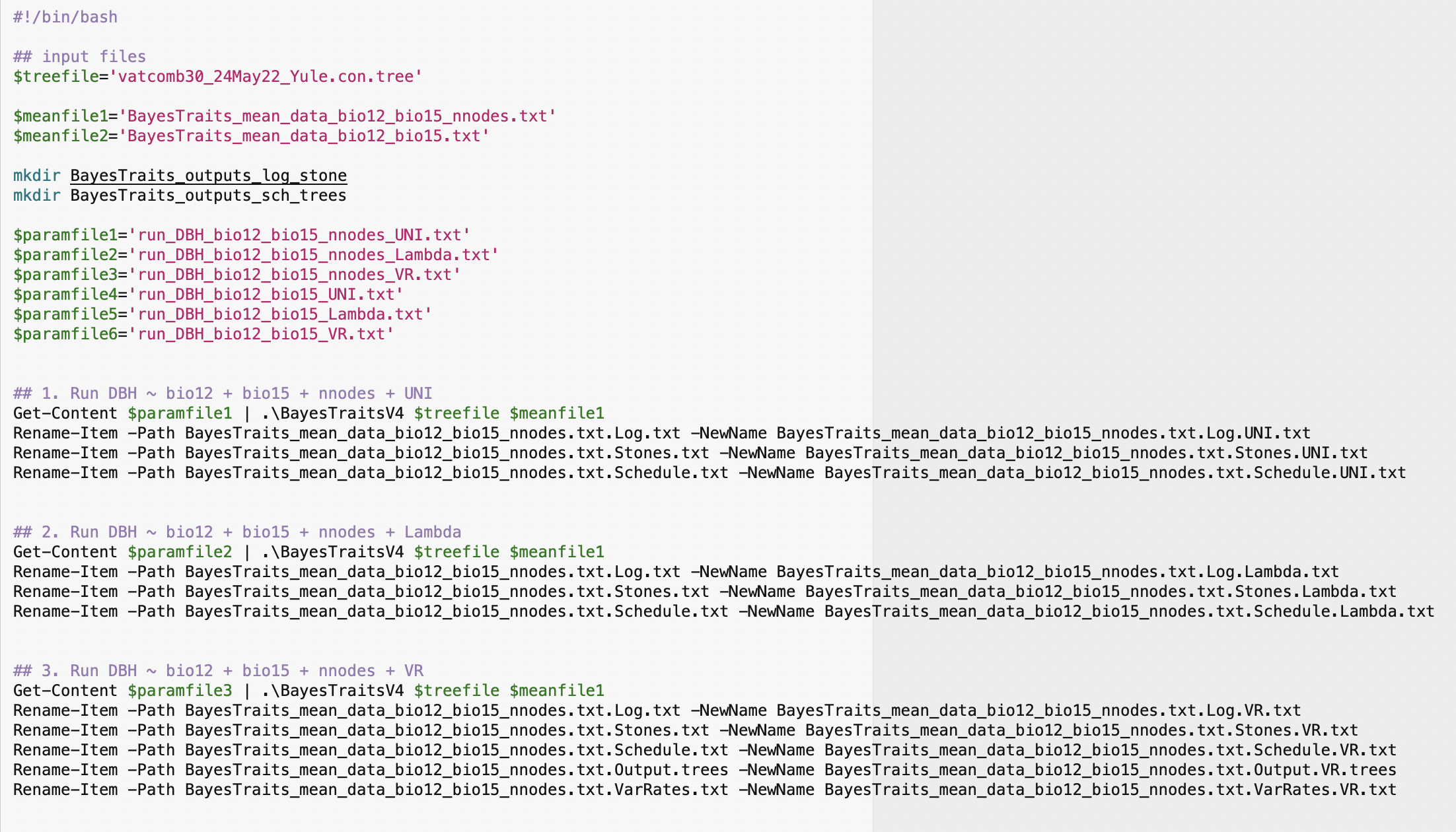
The function generates all running BayesTraits commands for the multiple regressions, depending on the parameters set in the arguments treetransf, bi, it, sa, and st.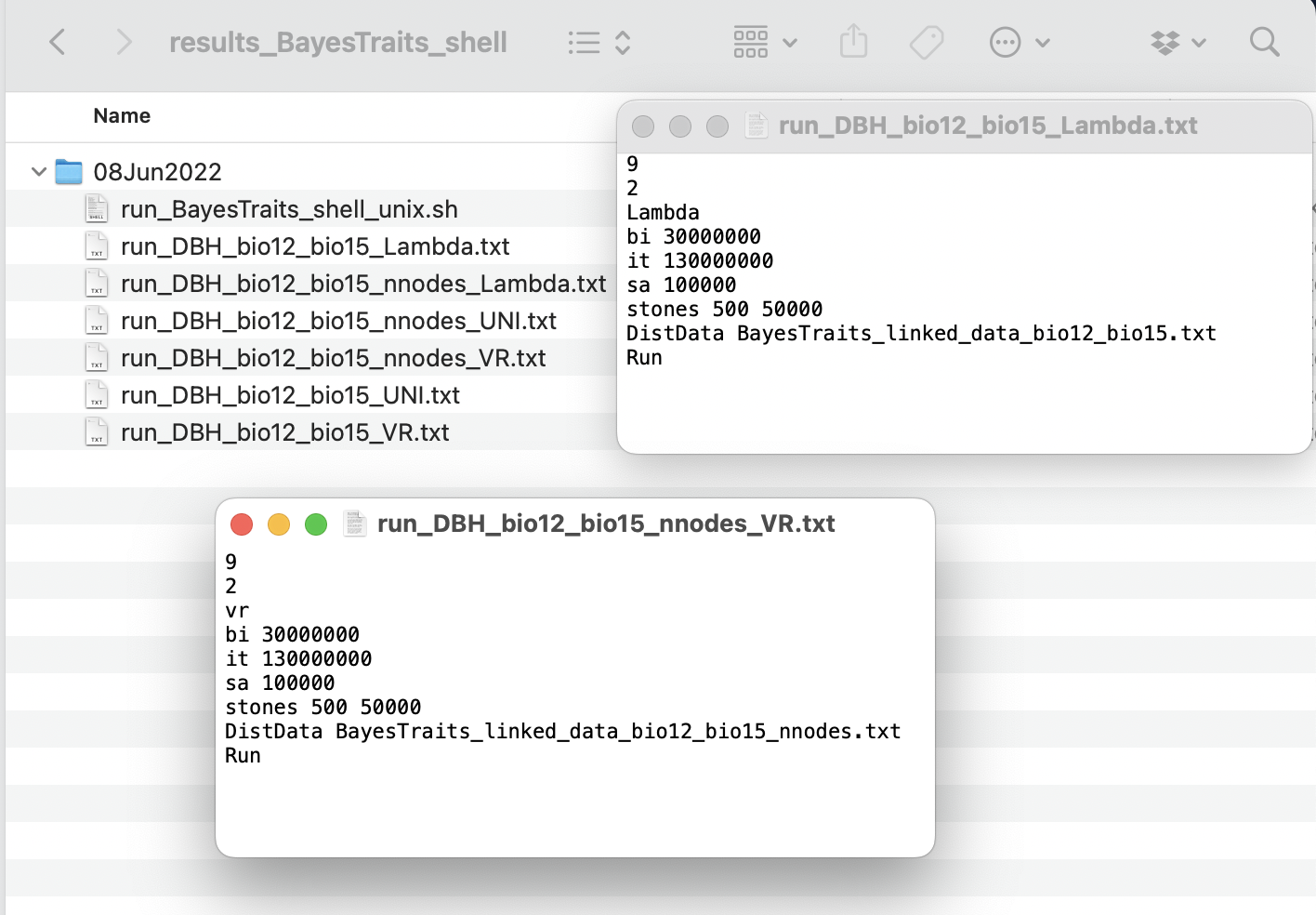
Note that because we have set the argument cc_DataTree = TRUE, then the tree, mean data, and linked data files were also copied directly into the BayesTraits folder.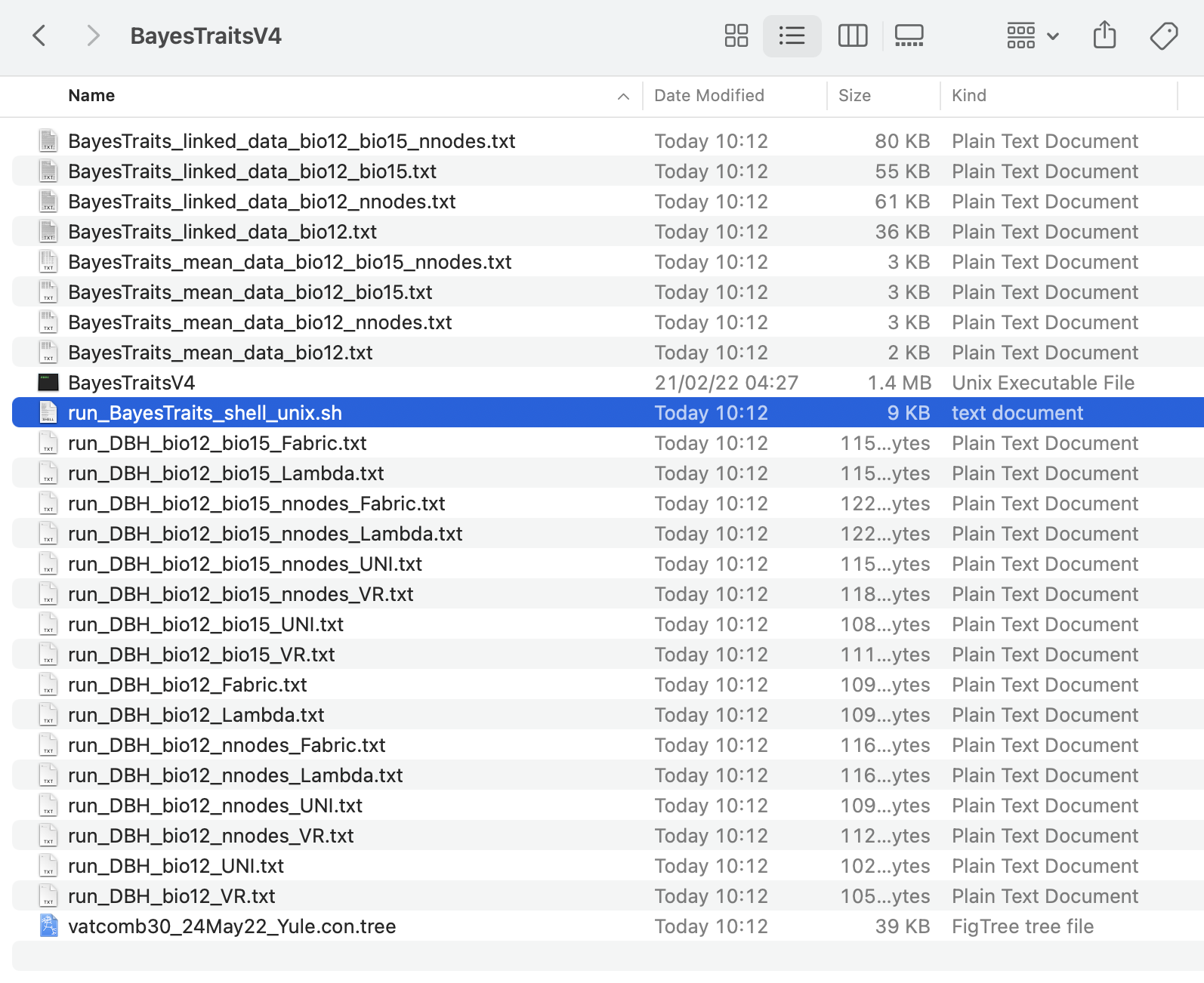
Note that if you set any of the arguments Kappa, Delta, Lambda, and OU as the default NULL, these tree transformation models will be estimated, provided that any of them were also listed in the argument treetransf. But you can also set them as a vector with any specific value between 0 and 1. For example, if you want to run analyses with both specific and estimated values for Lambda, then you have to set a vector such as c(“0.001”, “1.0”, “Estimate”).
Also, if you define the logical argument VRLambda as TRUE, then the specific values set for the tree transformation models at the parameters Kappa, Delta, Lambda or OU will be considered as separate runs when using the model of variable rates VR, provided that you have also chosen this model in the parameter treetransf.
Running the Unix shell script
After using the function phyreg.shell with all the needed parameters, you have to set permission to the shell script to run automatically all the phylogenetic regression analyses with BayesTraits program. You can just follow the steps below.
1. Open the terminal;
2. Go to the BayesTraits program folder where you have saved the shell script file .sh and the associated running and input data files with the function phyreg.shell. The shell script should be named run_BayesTraits_phyreg_shell_unix.sh;
3. Execute the following line:
chmod 777 run_BayesTraits_phyreg_shell_unix.sh
4. Then perform the BayesTraits run using the shell script by executing the following line:
./run_BayesTraits_phyreg_shell_unix.sh
Running the Windows PowerShell script
After using the function BayesTraits.shell with all the needed parameters, you have to set permission to the PowerShell script to run automatically all the phylogenetic regression analyses with BayesTraits program. You can just follow the steps below, but you may also want to find further information on how to use Windows PowerShell.
1. Open PowerShell as Administrator;
2. Go to the BayesTraits program folder where you have saved the PowerShell script file .ps1 and the associated running and input data files with the function phyreg.shell. The PowerShell script should be named run_BayesTraits_phyreg_shell_windows.ps1;
3. Execute the following line and choose option [Y]Yes;
Set-ExecutionPolicy -ExecutionPolicy RemoteSigned -Scope LocalMachine
4. Execute the following line:
Unblock-File -Path .\run_BayesTraits_phyreg_shell_windows.ps1
5. Then perform the BayesTraits run using the PowerShell script by executing the following line:
.\run_BayesTraits_phyreg_shell_windows.ps1
See also an associated article on how to automatically create input files with phyreg.inputs for the phylogenetic regression analyses, as well as another article on how to use phyreg.outputs to process the resulting output files from the analyses.