3. Processing output data from independent contrast regression analysis
Source:vignettes/articles/phyreg_outputs.Rmd
phyreg_outputs.Rmd
The main goal of the InNOutBT package is to readily create input data and process output data for Meade & Pagel’s (2022) BayesTraits program.
Here we show how to use the package’s function BayesTraits.outputs for processing all Log and Stones output text files from BayesTraits phylogenetic independent contrast regression analyses.
Setup
InNOutBT is not on CRAN yet but you can install the latest development version from GitHub:
#install.packages("devtools")
devtools::install_github("DBOSlab/InNOutBT")Processing Log and Stone output files
The function BayesTraits.outputs reads and processes the Log and Stones .txt output files resulting from the independent contrast regression analyses and generates tables in Word or CSV formats, which summarizes competing model and ranks them by log likelihood scores. Heatmaps derived from pairwise Bayes Factors are generated in PDF format and these can be used to identify competing modes that are most similar to the model with the highest log likelihood score.
library(InNOutBT)
phyreg.outputs(logst_dir = paste0(getwd(),"/BayesTraits_phyreg_outputs_log_stone"),
responvar = "DBH",
explanvar = c("bio12", "bio15", "nnodes"),
treetransf = c("Kappa", "Delta", "Lambda", "OU", "UNI", "VR", "Fabric"),
bayesfactor = TRUE,
unirates = FALSE,
outformat = c("Word", "CSV"),
tableleg = "Phylogenetic regression in BayesTraits",
dir_create = "results_BayesTraits_phyreg_output",
outfile = "BayesTraits_phyreg_output_table",
height = 12,
width = 15)
Heatmap including Bayes Factor pairwise comparison: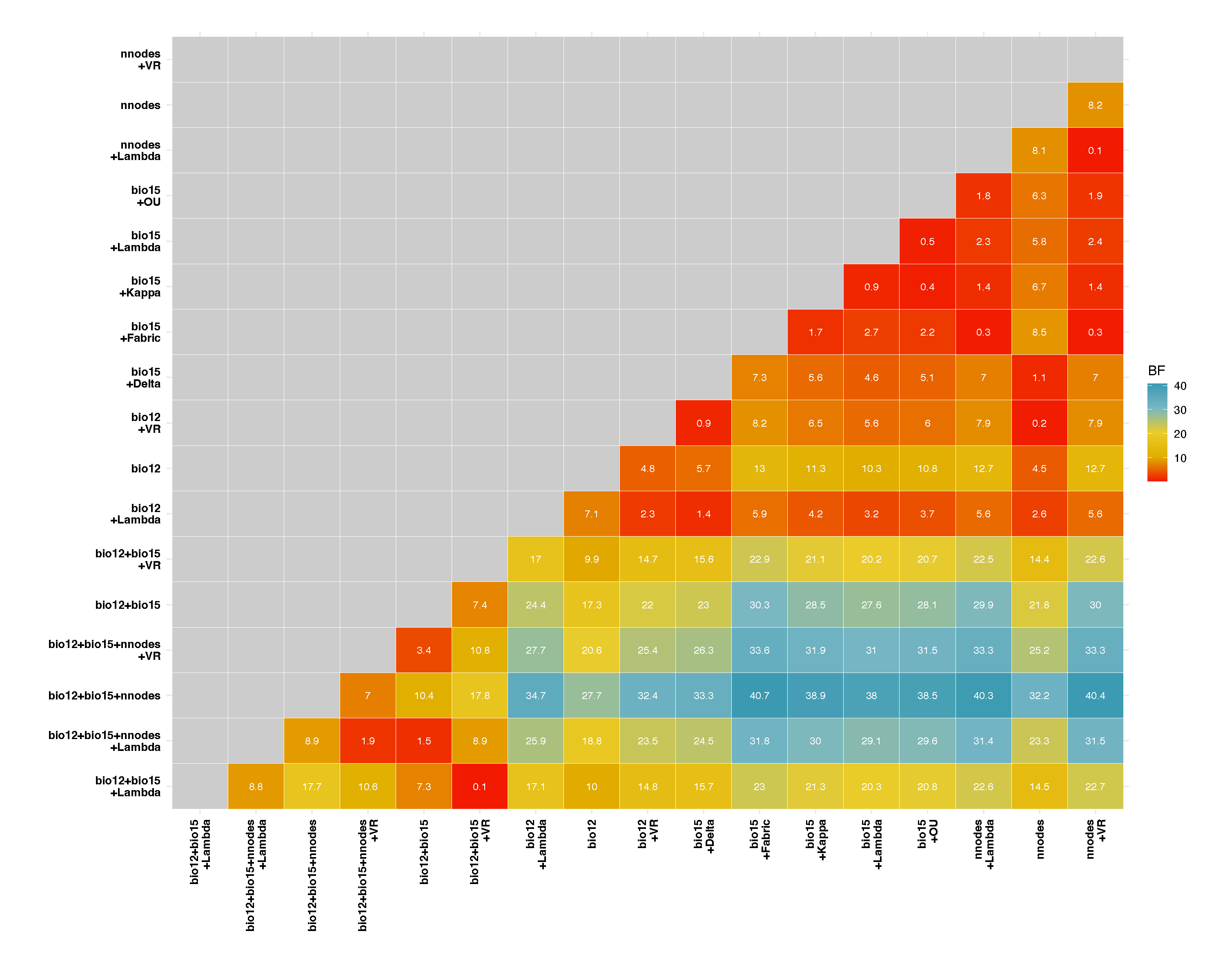
Heatmap clustering models based on Bayes Factor values: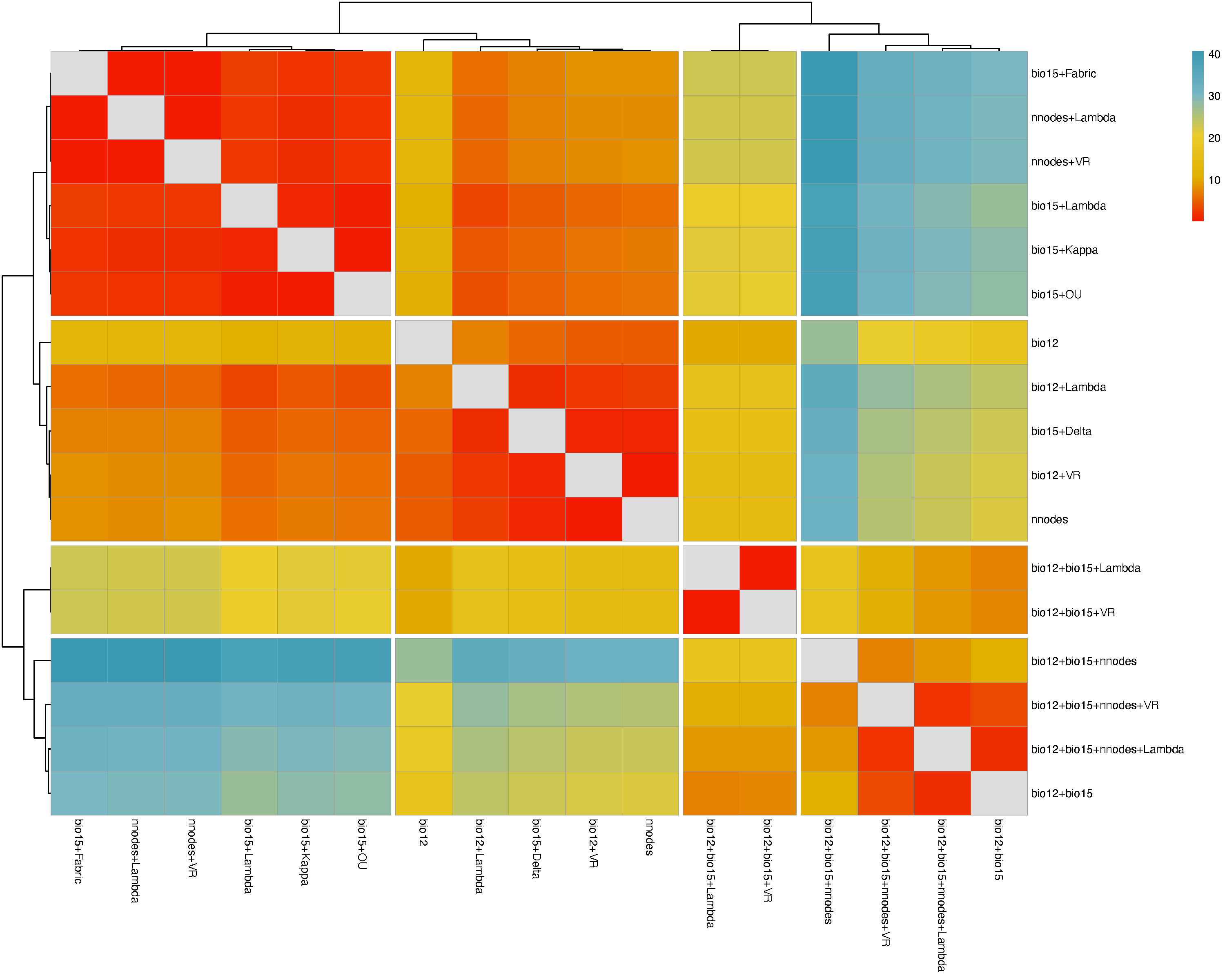
Table describing the models and coefficients: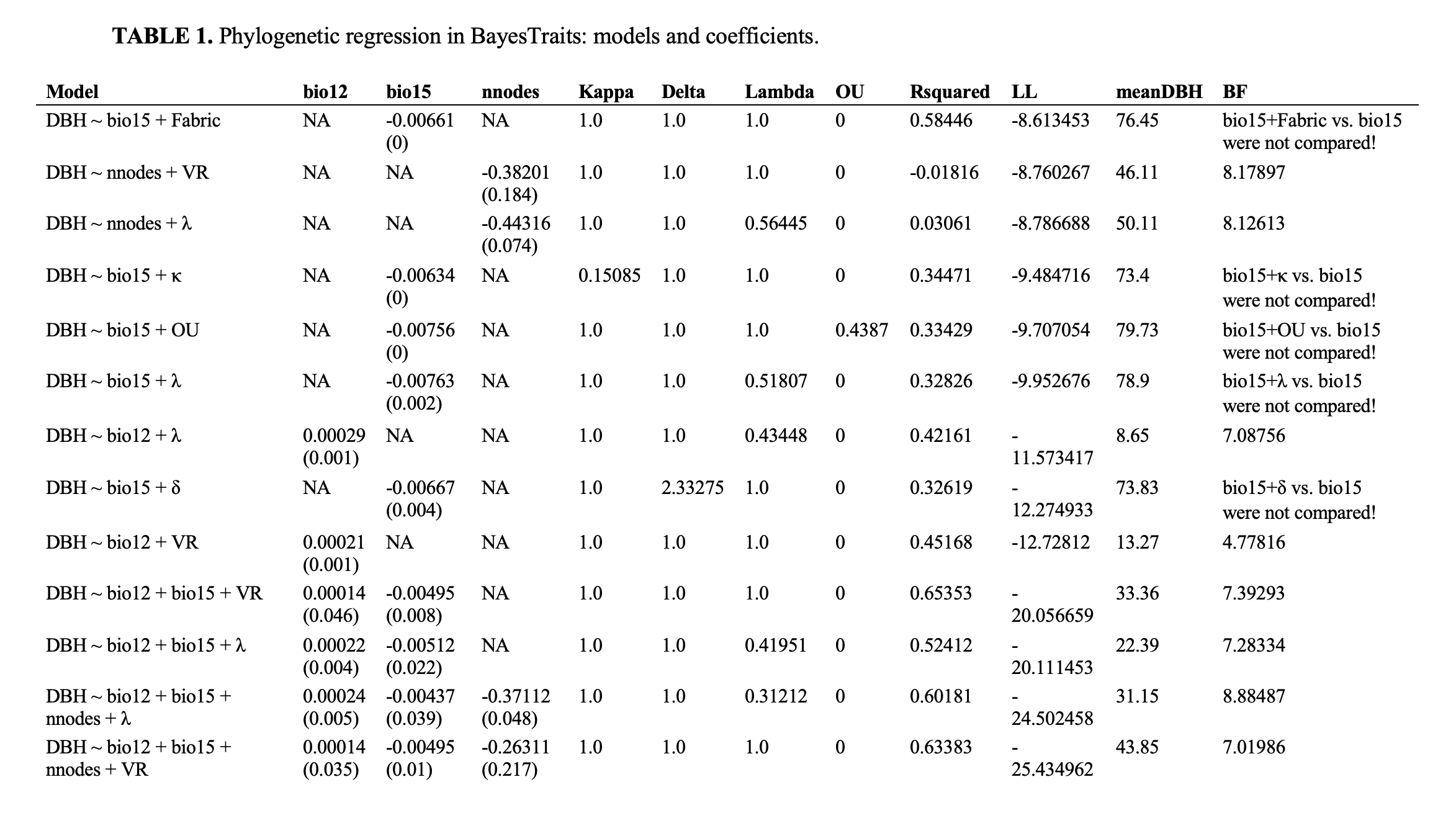
See also associated article on how to create input files with phyreg.inputs for independent contrast regression analyses, as well as another article on how to create a shell script with phyreg.shell for the analysis of one or multiple models.